Active Learning on Graph Nets
tags: [gnn gaussian_process machine_learning Graph networks have become very popular for representing and modelling materials and molecular structures. Like most deep learning approaches GNNs tend to require rather large training sets (minimum a few thousand examples to outperform classical ML approaches), another issue is the lack of confidence intervals on predictions. In this work we set out a method to address both of these problems.
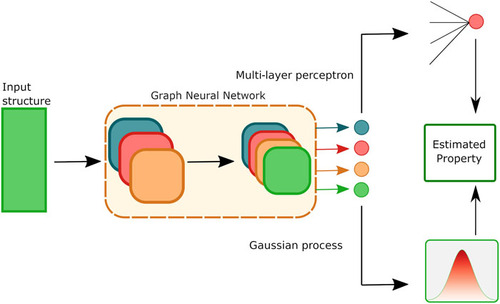
We use a GNN as a representation learner, producing a compact vector representation encoding information about the crystal structure and composition. This encoding is then used to train a Gaussian process, a class of Bayesian inference. The GP naturally provides a predicion and a variance, thus providing a confidence interval.
The confidences can then be used to develop an active learning scheme. In AL we train on a limited set of labelled data and then run inference on a larger set of un-labelled data. Using the uncertainty from the GP we can choose the next best data point for which to obtain a label, points with higher uncertainty will provide more information to the model for training. We demonstrate that model improvement is more than twice as efficient with new data points compared to random sampling when using the GP-based approach.